We’re excited to release GeoFlow V2, the state-of-the-art protein foundation model unifying structure prediction and de novo design at the atomic-level, providing for the first time a unified framework for understanding, discovering and optimizing the molecules of life.
- Try GeoFlow V2 for free now: https://prot.design (patent restrictions apply)
- Technical report: https://prot.design/report
A unified, all-atom diffusion model for protein understanding and generation
The 2024 Nobel Prize in Chemistry was awarded to key breakthroughs in protein structure prediction and design, namely AlphaFold and RFDiffusion. GeoFlow V2 now unifies prediction and generation in a single model, offering higher accuracy, designability, and applicability, paving the way for more ground-breaking breakthroughs in industrial protein design. Scientists can now generate proteins de novo targeting specific epitopes on multiple types of biomolecules, including other proteins, small molecules, DNA, and RNA.
Unlike most existing models that either predict or generate, GeoFlow V2 does both in a single model — accurately predicting the structures for provided inputs while designing new structures for masked tokens.
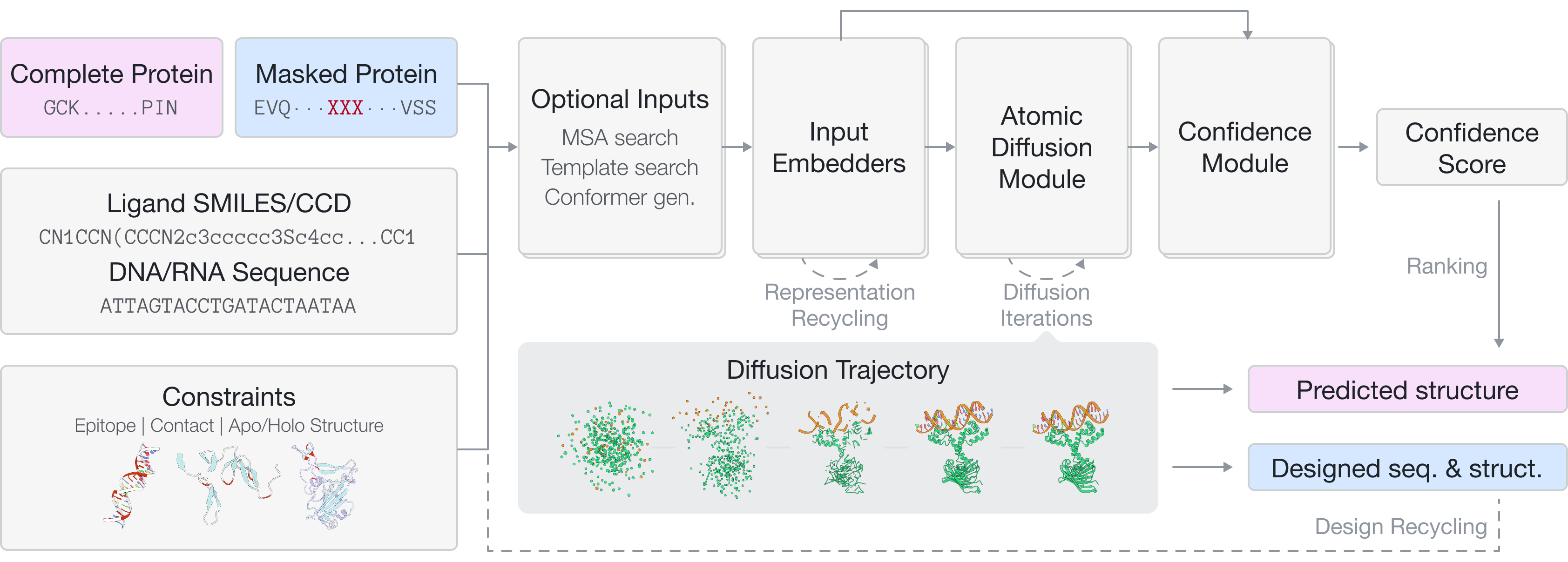
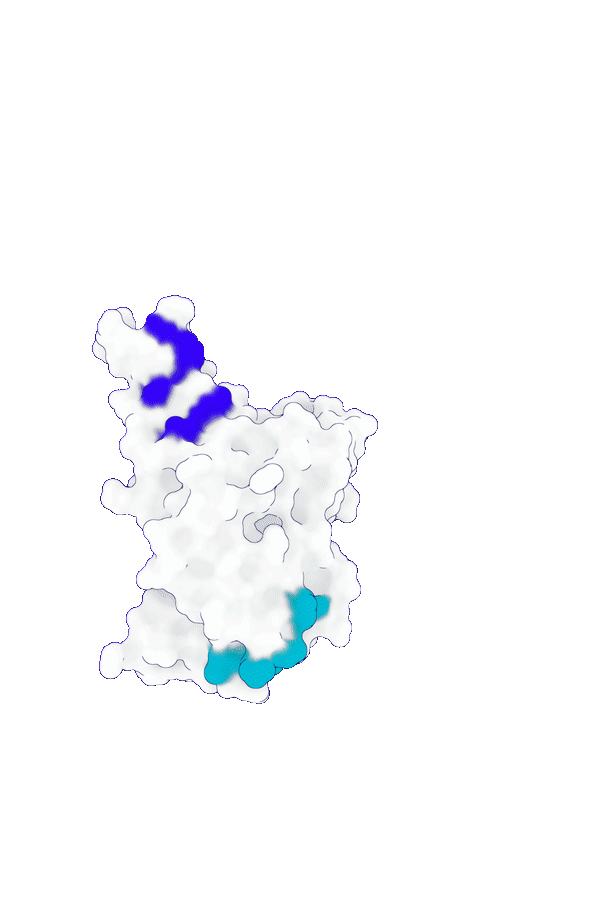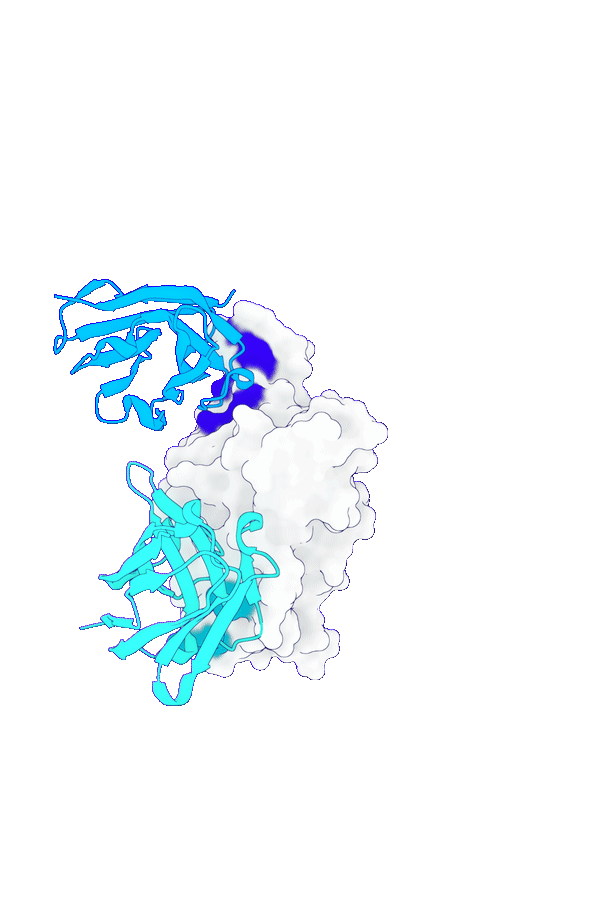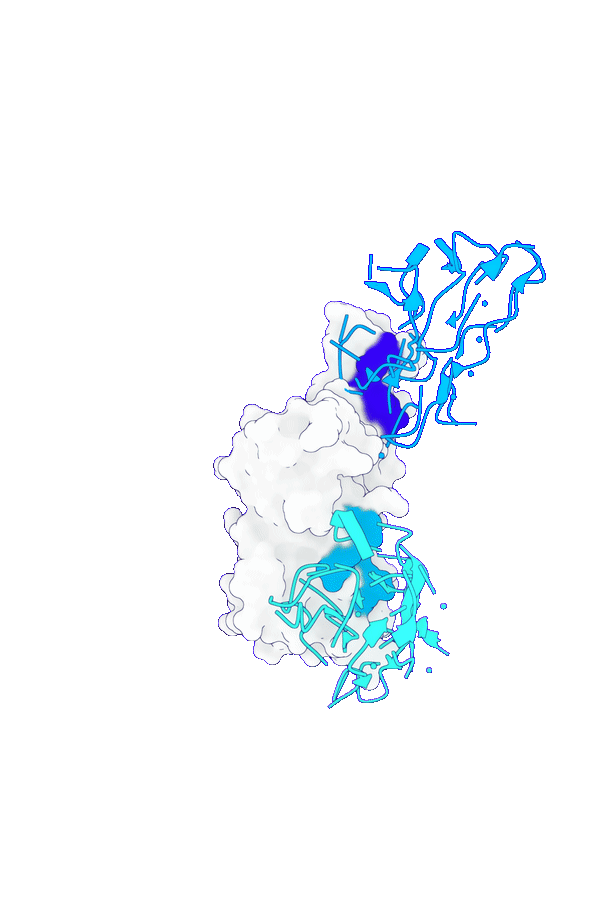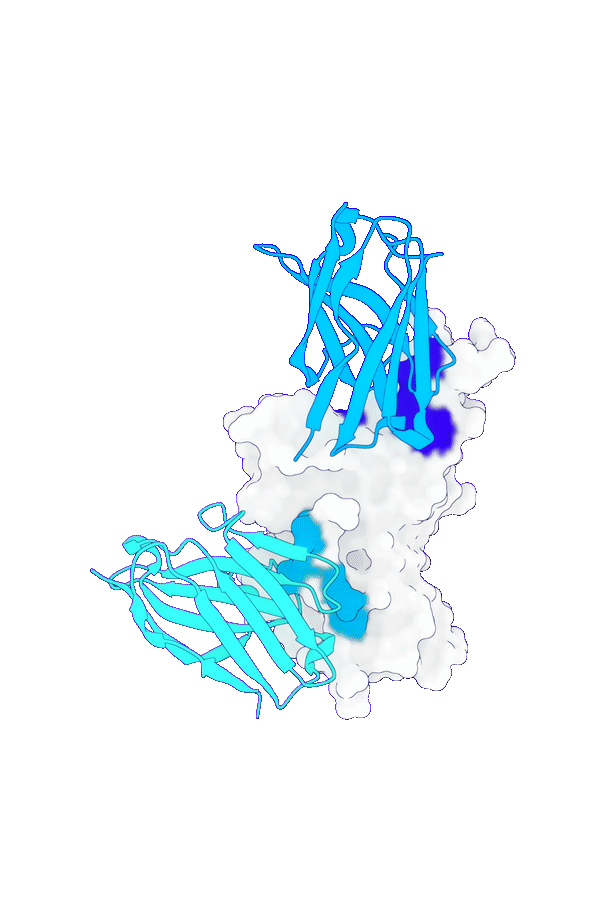
Accurate structure modeling for complex biomolecular systems
GeoFlow V2 excels in modeling biomolecular interactions under complex systems such as antibody:antigen, protein:DNA and protein-ligand. Specifically, for low-homology antibody-antigen structure prediction, GeoFlow V2 achieves a Top-1 success rate of 45% (DockQ > 0.23), significantly outperforming leading structure prediction models like Chai-1 (33%), Protenix (31%), and AFM2.3 (25%).
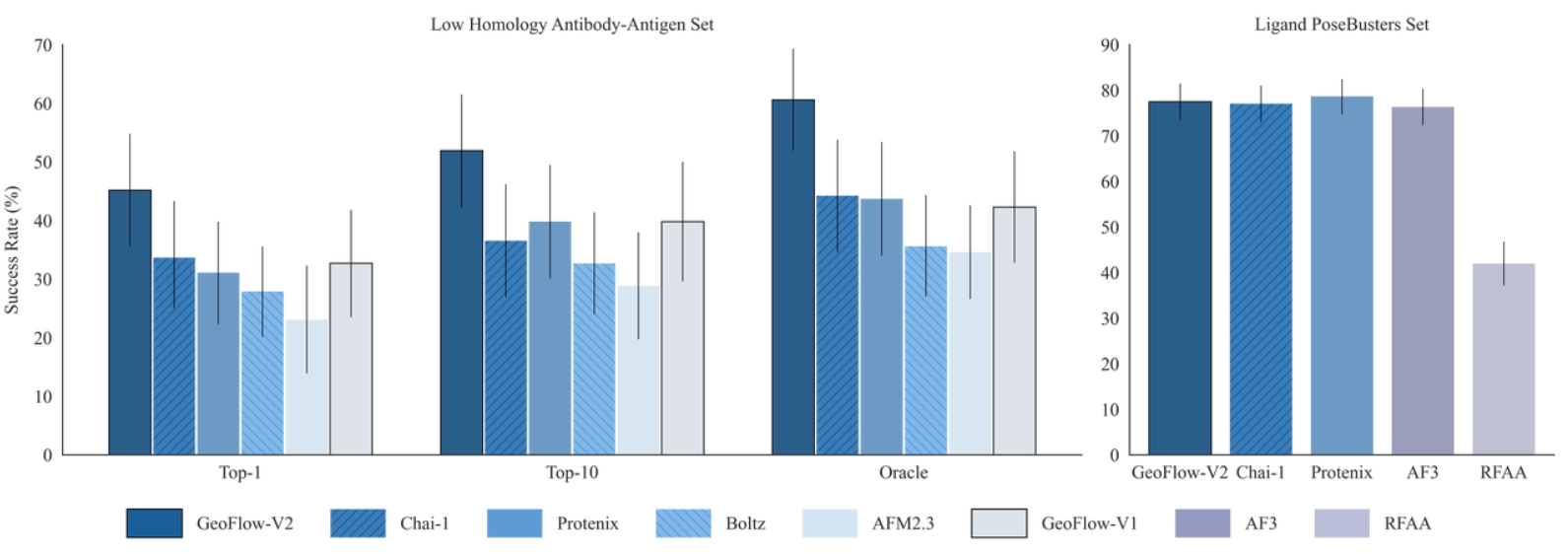
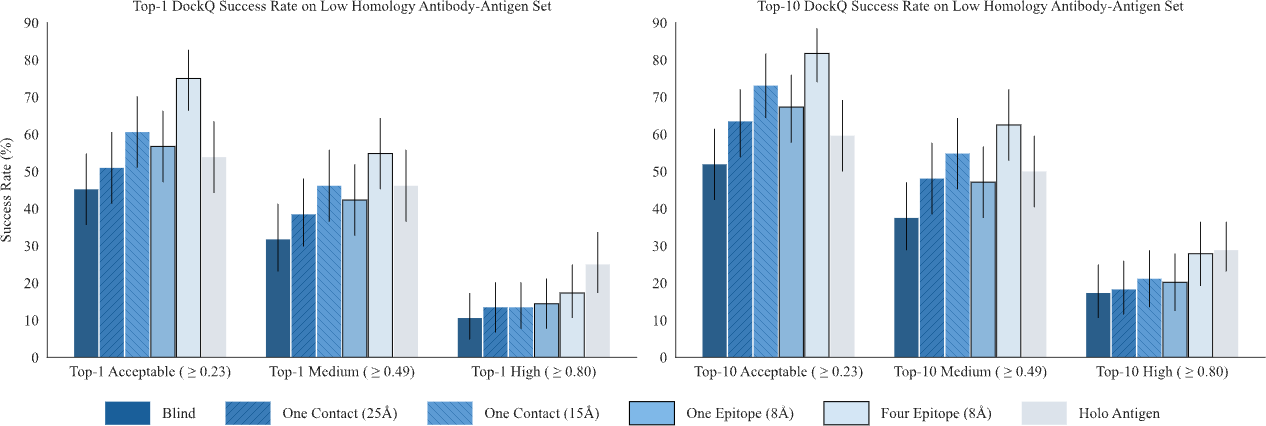
The structure prediction performance can be further boosted by adding various structural cues, including hotspots (token), epitopes (token-to-chain), contacts (token-to-token), and structural templates. The success rate increases to 75% when four accurate epitope hints are provided, demonstrating GeoFlow V2’s precise modeling of atomic-level interactions, a crucial ability for epitope-based antibody design.
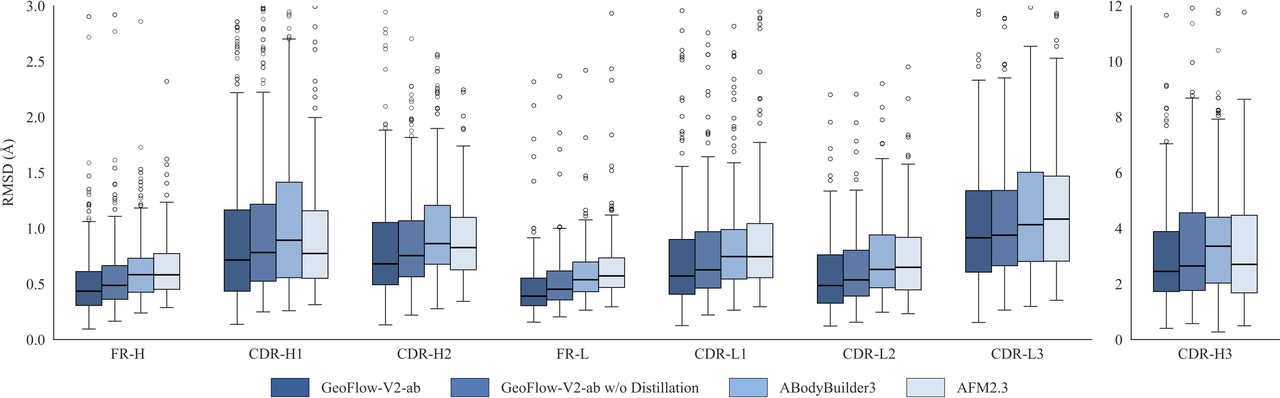
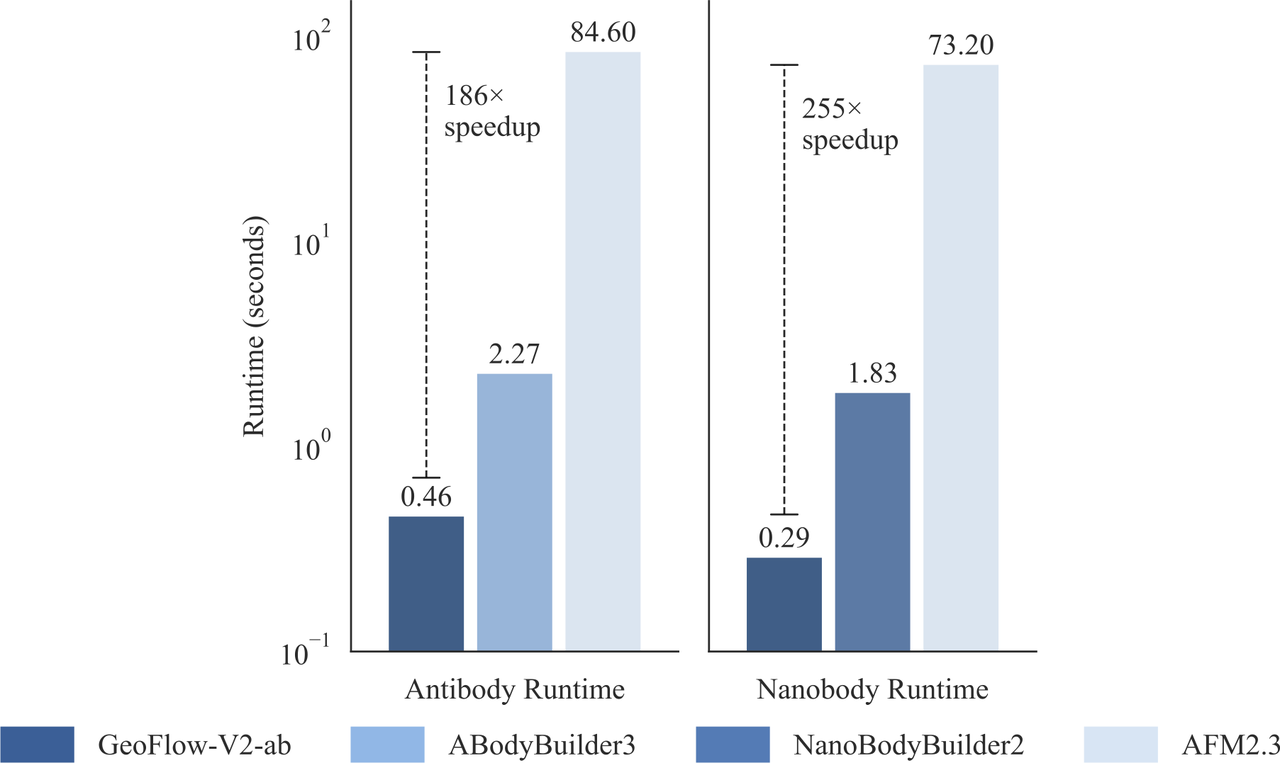
In addition to the large-scale foundation model, a smaller distilled model GeoFlow V2-ab was trained for lightning-fast antibody structure prediction. GeoFlow V2-ab runs over 100 times faster than AFM2.3 while being more accurate. The subsecond-level inference time enables fast inference and virtual screening of millions of antibody structures in a single campaign.
Towards high-quality, controllable in silico protein design
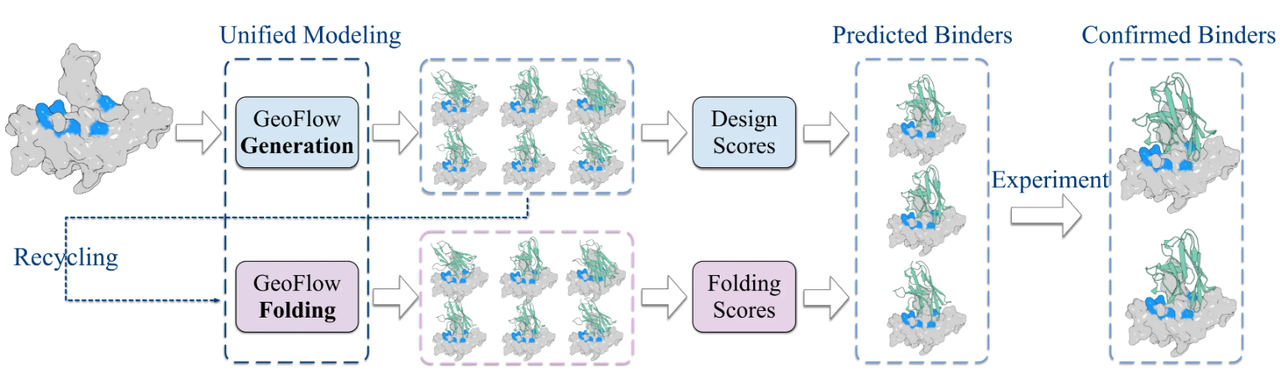
Designing novel, stable and functional protein has numerous applications in biomedicine (e.g. antibodies / vaccines), and synthetic biology (e.g. enzymes). By unifying design and prediction at the atomic level, GeoFlow V2 can easily be applied to mass-scale, highly-specific protein design campaigns, where it first generates the target:binder complex sequence and structure from an incomplete complex, and then re-folds the complete sequence into the complex structure to calculate self-consistency. The highest-ranked candidates are then synthesized and validated in wet lab.
In antibody design benchmarks, GeoFlow V2 demonstrates industry-leading epitope compliance, CDR contact rate, and virtual screening pass rate compared to the previous state-of-the-art RFAntibody (fine-tuned from RFDiffusion on antibody datasets).
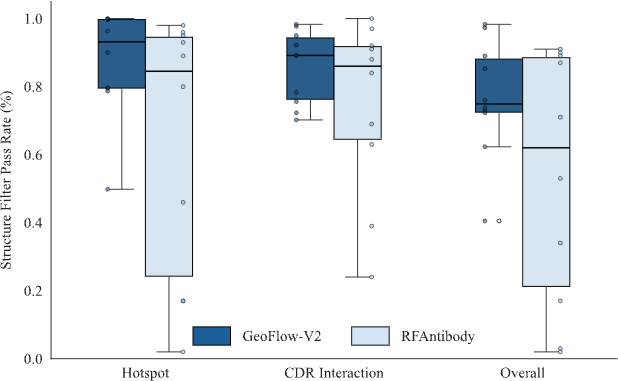
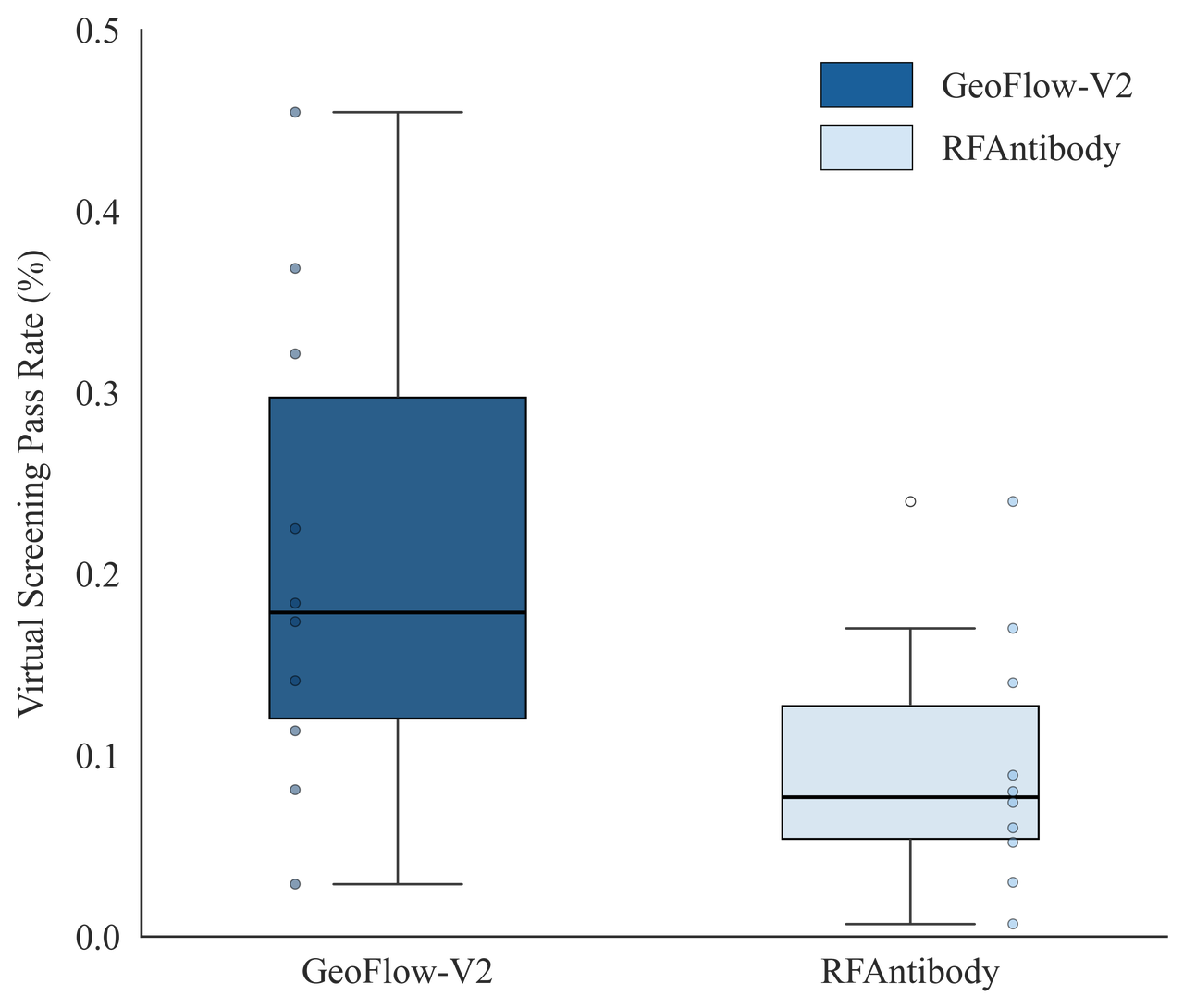
In an antibody design benchmark composed of seven targets, 1,000 epitope-specific antibodies were generated with framework region fixed to the Trastuzumab framework. GeoFlow V2 successfully recovered the reference Ag:Ab binding mode on 5 out of 7 targets (minimum FR RMSD < 8Å), while RFAntibody succeeded on 2/7 targets.
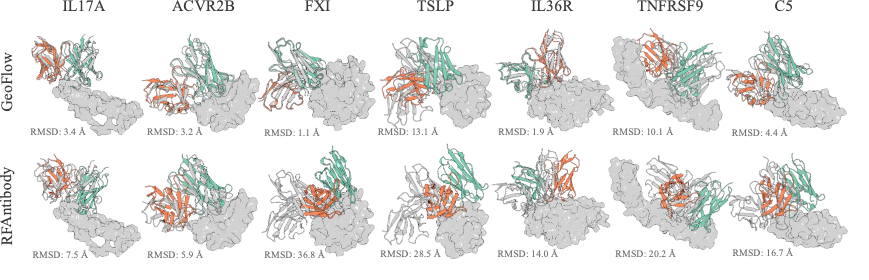
The above results indicates a potentially higher success rate for de novo antibody design given the same wet-lab budget. Stay tuned for our updates in wet-lab results!
Real-world applications of GeoFlow V2
Traditional protein discovery relied on the combination of complex systems evolved in nature (e.g., using animal immune systems to discover antibodies), mass-scale trial-and-error (e.g., saturation mutagenesis), or expert experience (e.g. rational design). GeoFlow V2 pushes protein design further into the age of AI, making it more precise, more applicable, and more scalable. Below are a few real-world examples.
1. Designing soluble analogs for transmembrane proteins
In partnership with a leading pharmaceutical company, GeoFlow V2 successfully designed a soluble analog of a multi-pass transmembrane protein. The model redesigned its hydrophobic transmembrane (TM) regions, creating soluble variants while maintaining its extracellular structure and function. Among the sequences designed by GeoFlow V2, 90% are thermostable and is capable of soluble expression, with several candidates matching the binding specificity of the wild-type molecule while having a merely 22% TM sequence similarity. Such analogs can be highly useful for binder screening against a specific antigen conformation.
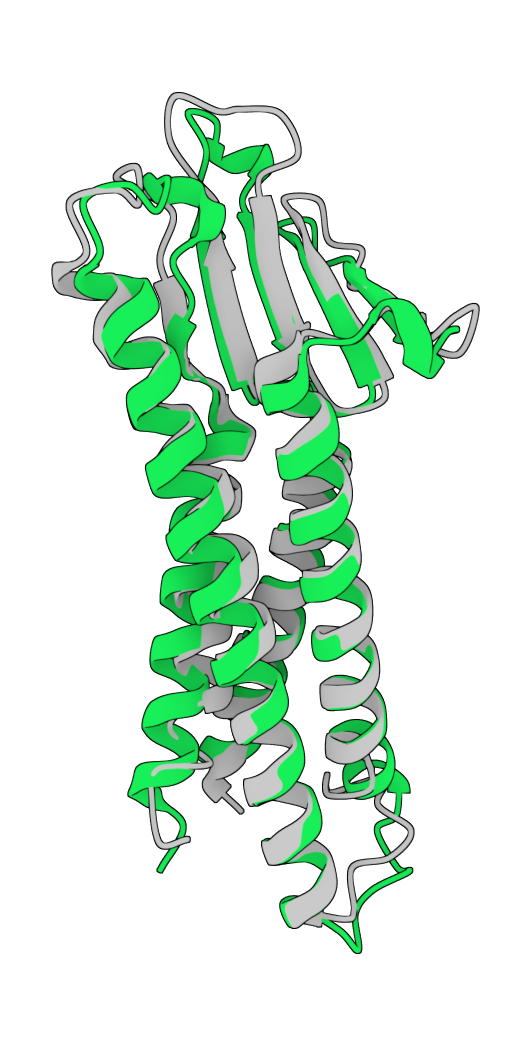
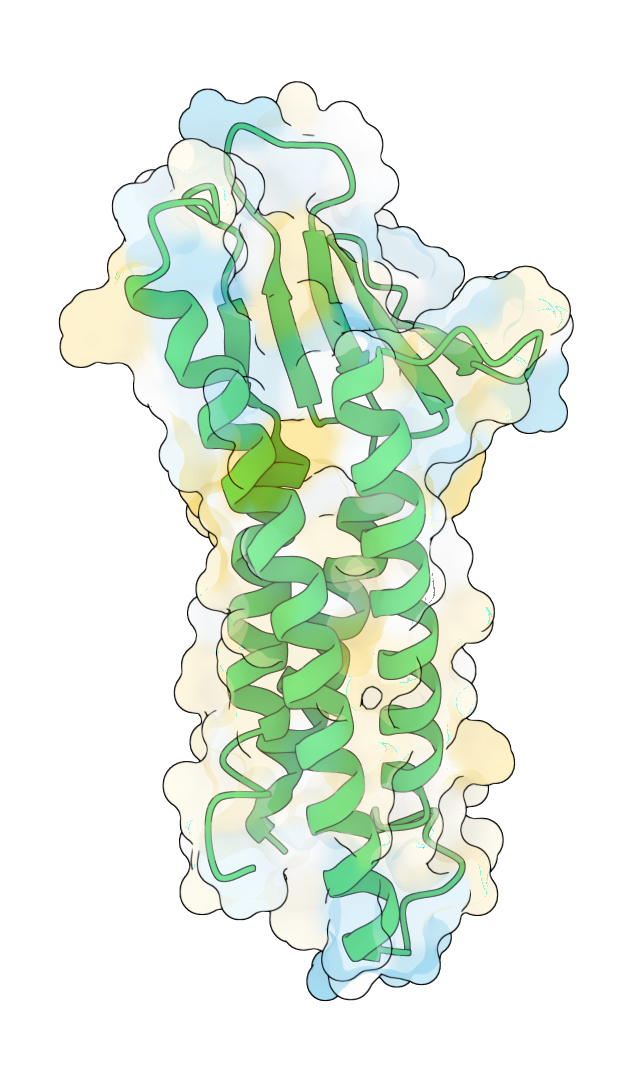
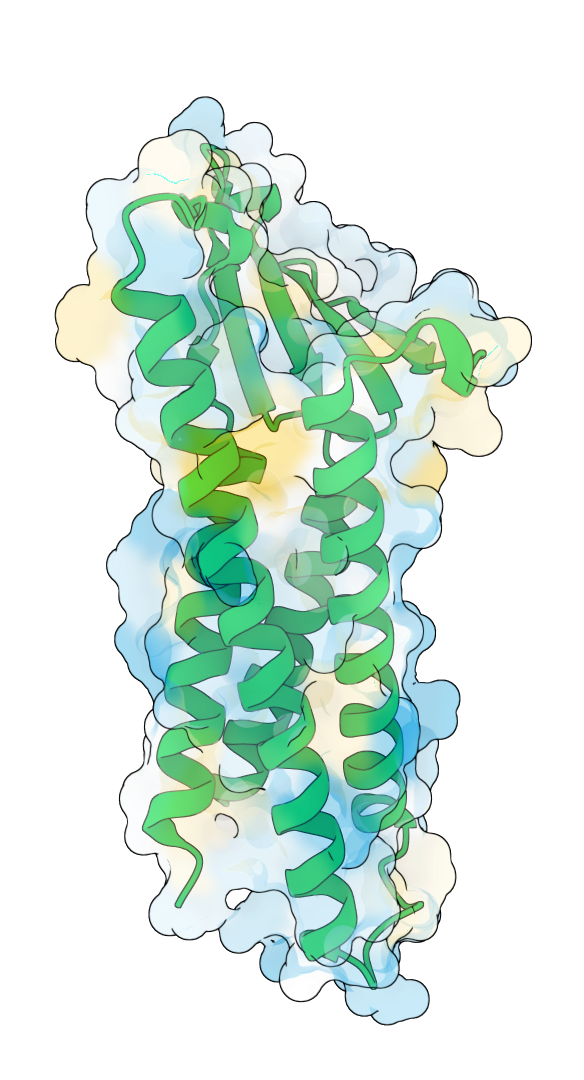
After design: hydrophilic TM region
2. De novo antibody design: targeting the previously undruggable
In several antibody design campaigns, multiple low-nanomolar antibody/nanobody candidates were identified after GeoFlow de novo design and high-throughput screening. Several antibodies have completed in vivo functional validation in animals. This new approach provides an alternative antibody discovery route in addition to animal immunization, opening new paths for highly specific antibody discovery against hard-to-drug epitopes.
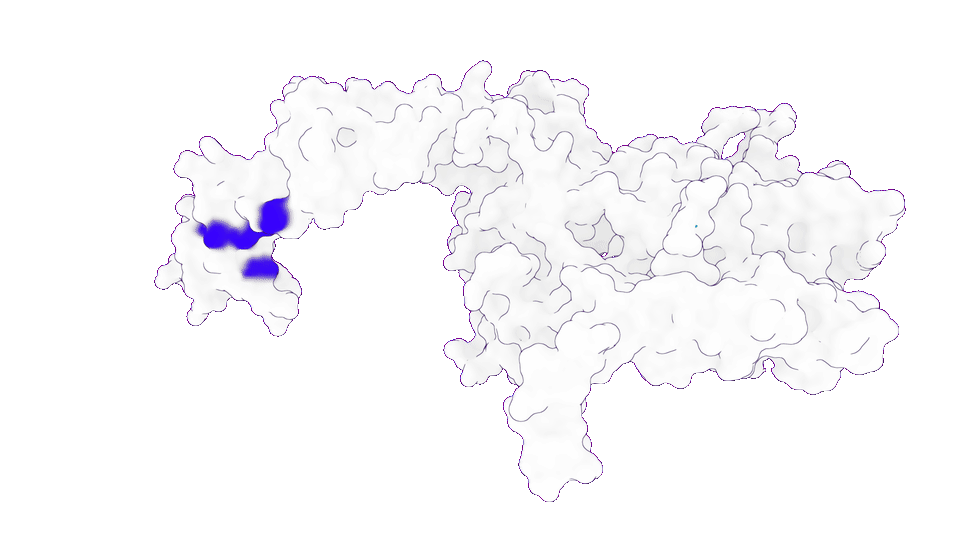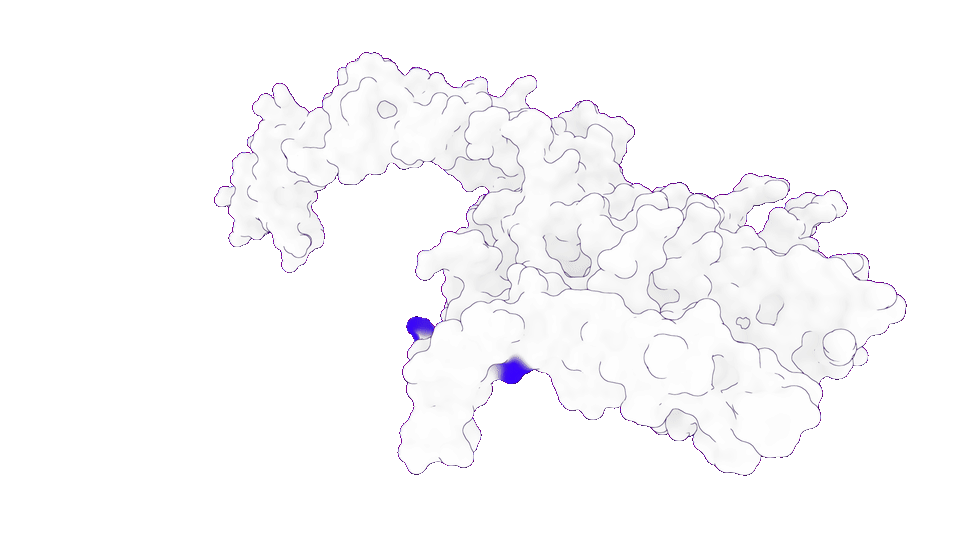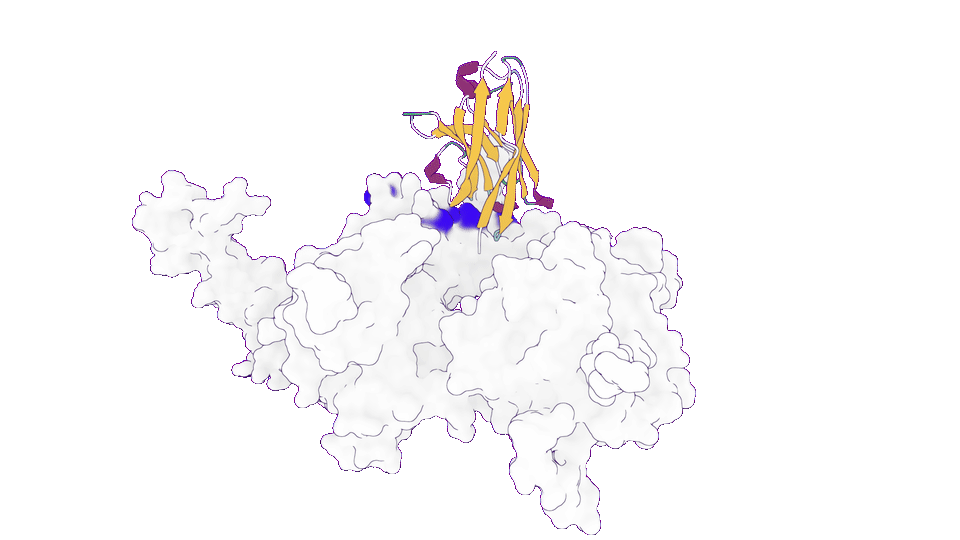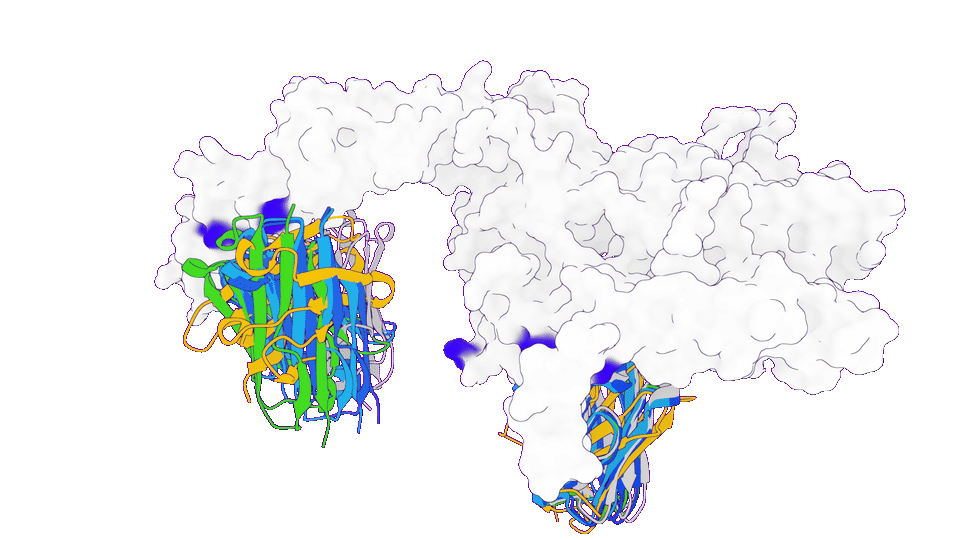
3. Obtaining patentable functional proteins at turbo speed
In a functional protein design project, GeoFlow V2 was tasked to redesign the target-binding loop to produce patentable, functional protein variants. Among the 100 candidate molecules generated in the first round, 80% expressed successfully with good thermostability, among which 50% has higher target-binding affinity (up to ~100 pM) than benchmark, which was obtained after 5 rounds of library construction and screening.
This showcased the potential of GeoFlow V2 to greatly shorten the development cycle for valuable protein products.
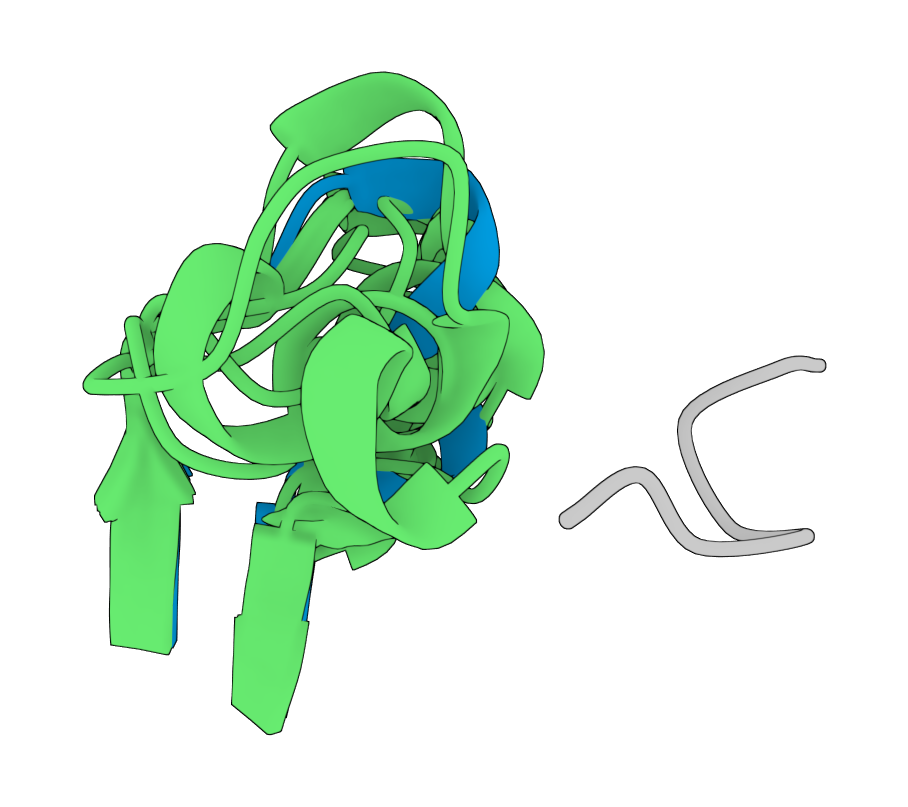
4. Achieving multi-objective optimization of Transaminase in 55 days
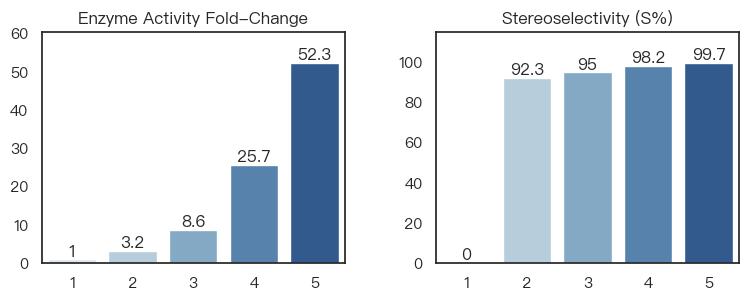
During the synthesis process optimization of a key pharmaceutical intermediate, GeoFlow V2 was employed for directed evolution of transaminases to address the catalytic challenge of a crucial chiral amine precursor. Through extensive interaction modeling and variant design, and leveraging an high-throughput, in-house experimental platform, BioGeometry achieved comprehensive enzyme performance optimization in just 55 days—boosting catalytic activity by 52.3-fold and increasing stereoselectivity for the (S)-isomer from 0% to 99.7%. All key metrics met the requirements for pilot-scale process development after the multi-objective optimization.
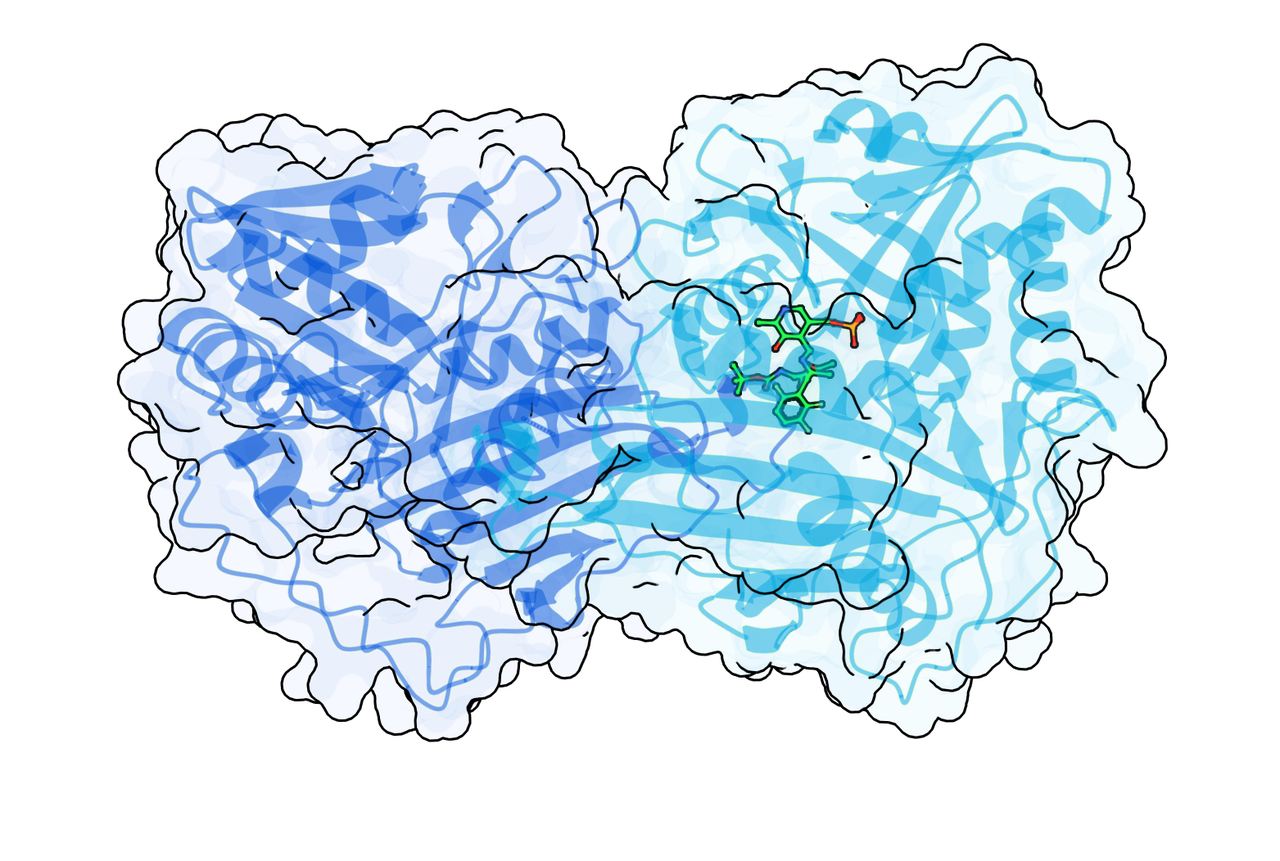
5. Designing a novel biosynthesis pathway with enzyme mining
A certain non-natural amino acid holds significant commercial value, but traditional synthesis routes have low yields below industry tolerence.
By using GeoFlow V2 for enzyme mining and 2-round optimization, BioGeometry has discovered a new enzyme capable of catalyzing a certain type of biochemical reaction. This enzyme enabled the design of a one-step in vitro biosynthesis route with strong commercial advantages (low cost, high yield).
GeoFlow V2’s unique powers were demonstrated through (1) the successful mining of the first reported enzyme for this type of biochemical reaction, and also (2) the improvement in its catalytic activity by 65 times over two rounds of enzyme optimization. Based on GeoFlow’s capabilities and BioGeometry’s in-house high-throughput wet lab, the entire enzyme development process was completed in just two months.
Infrastructure for the next-generation proteins, available now
Unifying structure prediction and generation into a single framework, GeoFlow V2 is taking one step further towards making life’s machine programmable. By understanding how nature writes its code, we are unleashing unprecedented productivity for next-generation protein design.
We know there are many steps to come, and we are committed to walking this path together with you. As from today, GeoFlow V2 is offering a limited-time free trial to researchers and engineers worldwide. Simply visit https://prot.design to get started. For deeper collaboration or custom services, please contact bd@biogeom.com.
