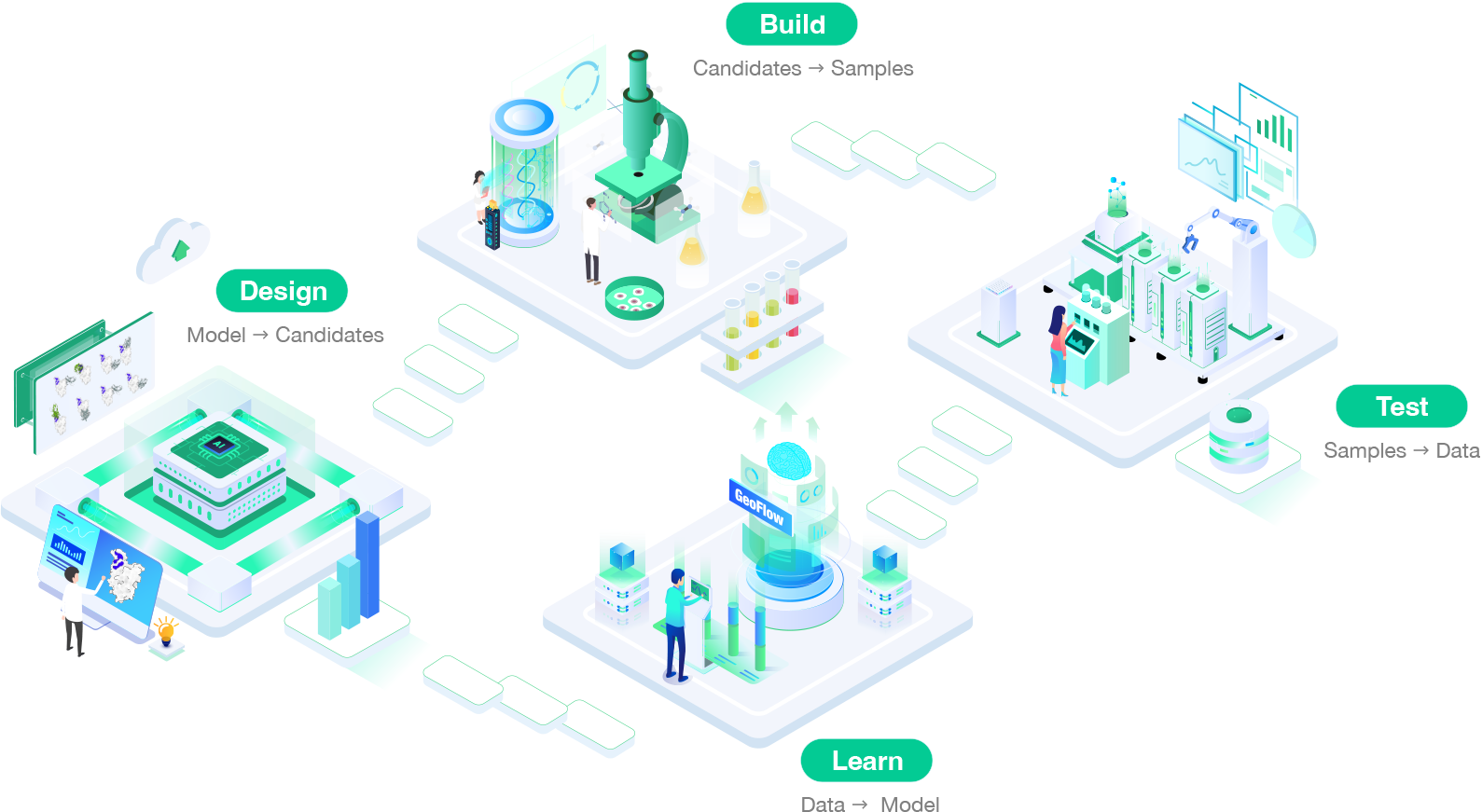
We combine AI with wet lab experiments to effectively reduce the volume of experiments, shorten the R&D cycle, and cut down the costs. In the cycle between dry lab and wet lab, it enhances the precision and confidence of our AI models, thereby increasing the clinical success rate of drugs.
Generative AI —Advancing Towards De Novo Protein Design
We are the first to apply generative diffusion models to molecular design. Deep generative models are expressive approximators for high-dimensional probability distributions. We have a repertoire of expressive generative models at hand for evaluating sample quality and generating novel samples.
Our Advantages:
- Expanded search space: Traditional rational design can optimize only a few amino acid sites at a time, while generative AI can design and optimize entire protein segments, such as the HCDR3 region.
- Customized generation: Design specific proteins according to binding targets/catalytic functions/property requirements; As far as imagination reaches, we strive to realize it for you.
- Rapid iteration: Quickly iterate models and molecules in high-throughput wet experiments to obtain the best molecules at the greatest speed.
Geometric Deep Learning – Understanding Proteins Based on Structure
Geometric deep learning is a cutting-edge deep learning technique that models proteins and other molecules in 3D space with the necessary symmetry in “mind”. We develop powerful GDL methods to dive deep into protein structures.
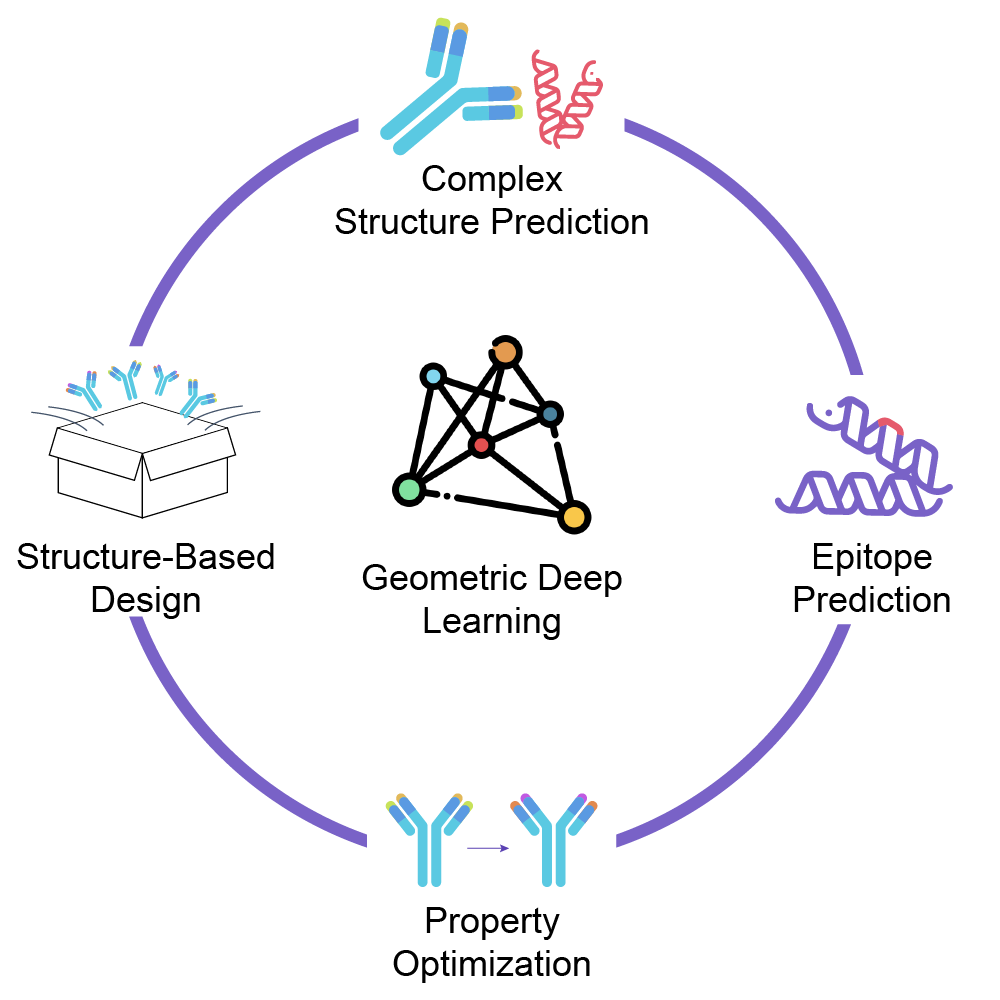
Our Advantages:
- Fast and accurate structure prediction: Our self-developed antibody structure prediction algorithm is 20 times faster than traditional methods with higher accuracy and smaller error.
- Insight into molecular binding modes: Accurate modeling of the binding interface enables effective de novo design and directed evolution of macromolecules.
- Anticipate risks and modify molecules: Based on the structure, various risks such as molecular stability and immunogenicity can be predicted. Coupled with generative AI for molecular modification, this can greatly increase the success rate of projects.
Selected Publications
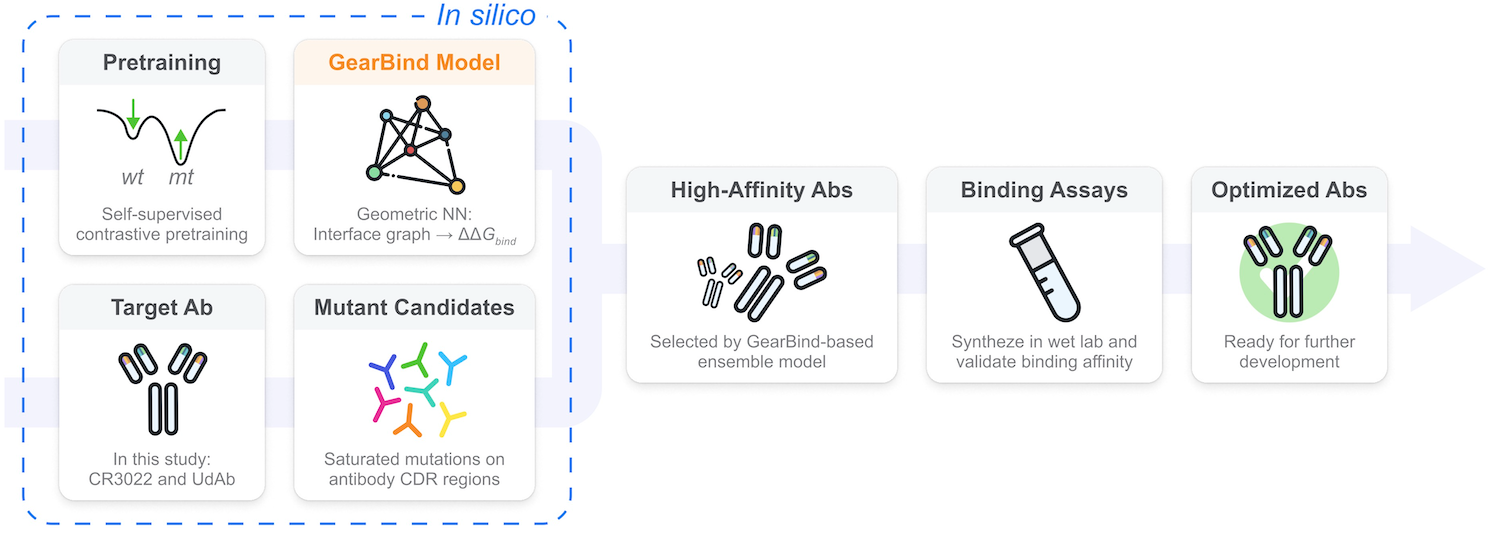
GearBind: Pretrainable geometric graph neural network for antibody affinity maturation
Geometric deep learning + contrastive pretraining yields 6x affinity in 20 samples.
Nature Communications, 2024
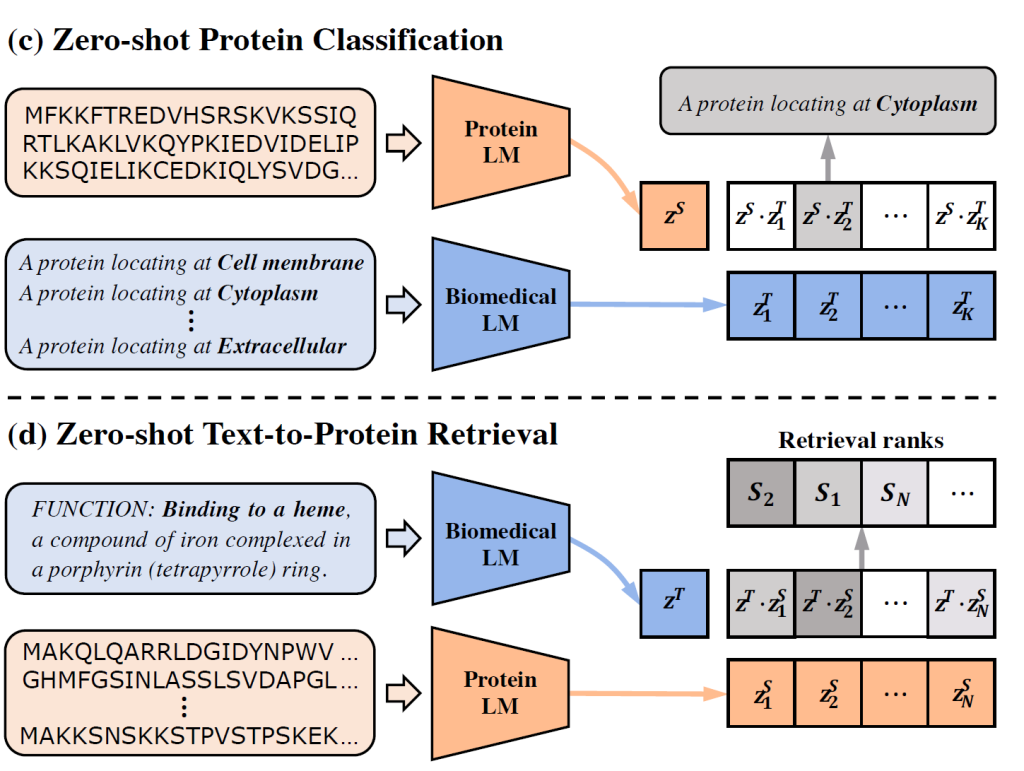
ProtST: Multi-Modality Learning of Protein Sequences and Biomedical Texts
Multimodal pretraining enables zero-shot function prediction and retrieval.
ICML 2023 Oral Presentation
ProtSeed: Protein Sequence and Structure Co-Design with Equivariant Translation
Joint sequence-structure translation enables fast generative protein design.
ICLR 2023
E3Bind: An End-to-End Equivariant Network for Protein-Ligand Docking
End-to-end protein-ligand docking with SE(3)-equivariance.
ICLR 2023
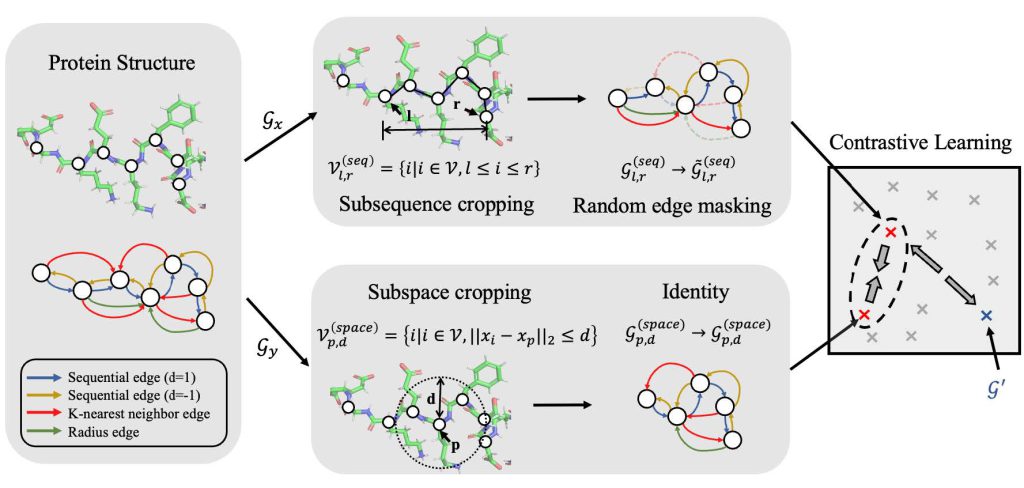
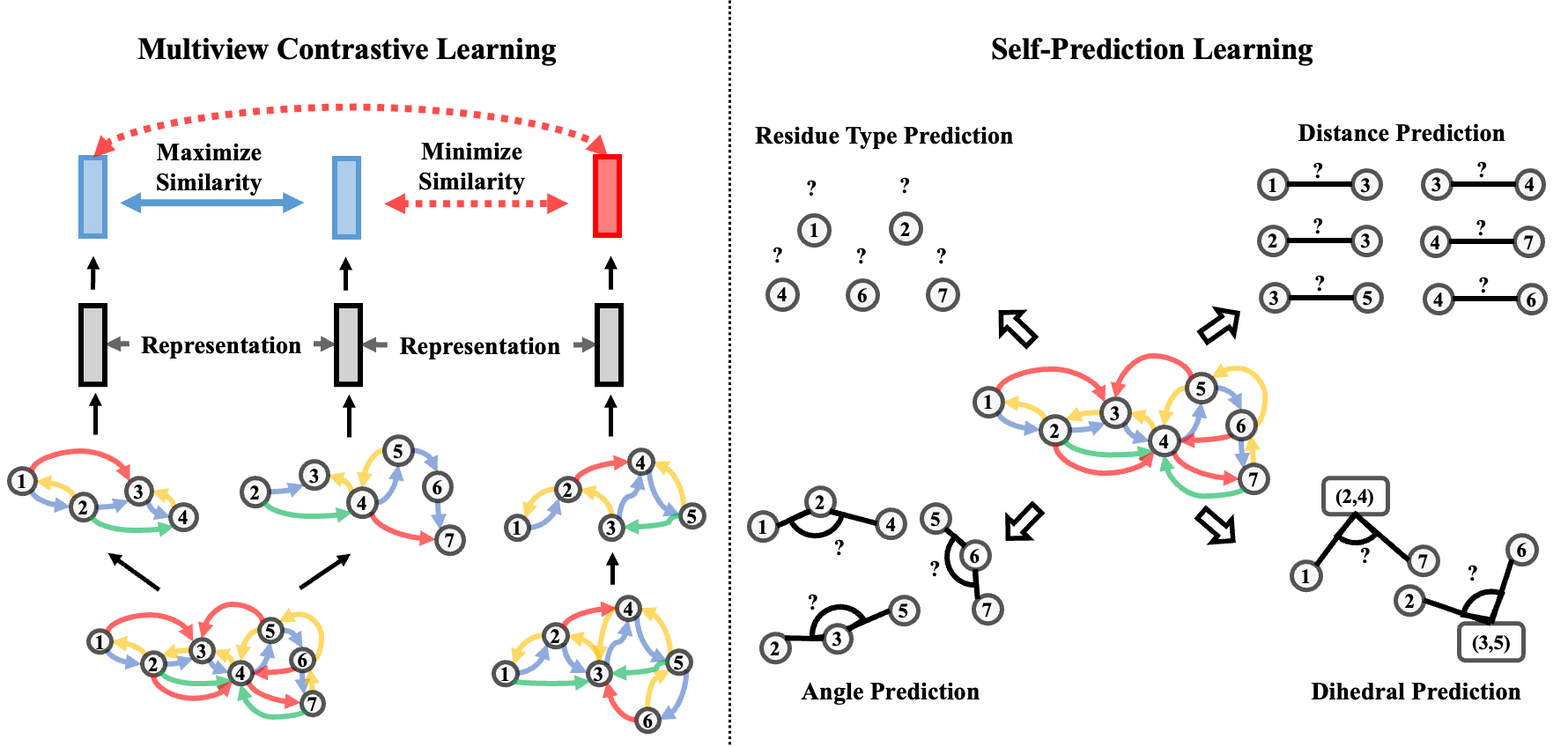
GearNet: Protein Representation Learning by Geometric Structure Pretraining
Multiview contrastive pretraining yields rich protein structure representations.
ICLR 2023
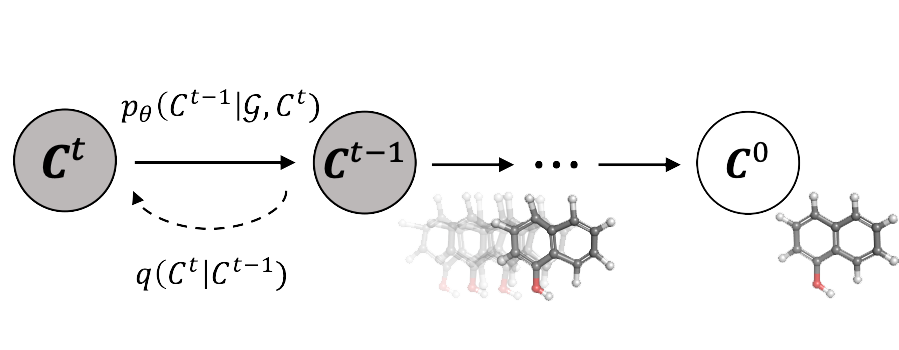
GeoDiff: A Geometric Diffusion Model for Molecular Conformation Generation
Geometric probabilistic models; Markov chains; SE(3)-equivariance; denoising diffusion.
ICLR 2022 Oral Presentation
PEER: A Comprehensive and Multi-Task Benchmark for Protein Sequence Understanding
Simple, effective structure-based protein encoder coupled with geometric self-supervised learning.
NeurIPS 2022 Datasets and Benchmarks Track

ConfGF: Learning Gradient Fields for Molecular Conformation Generation
Molecular 3D conformation generation; denoising score matching; SE(3)-equivariance.
ICML 2021 Long Talk
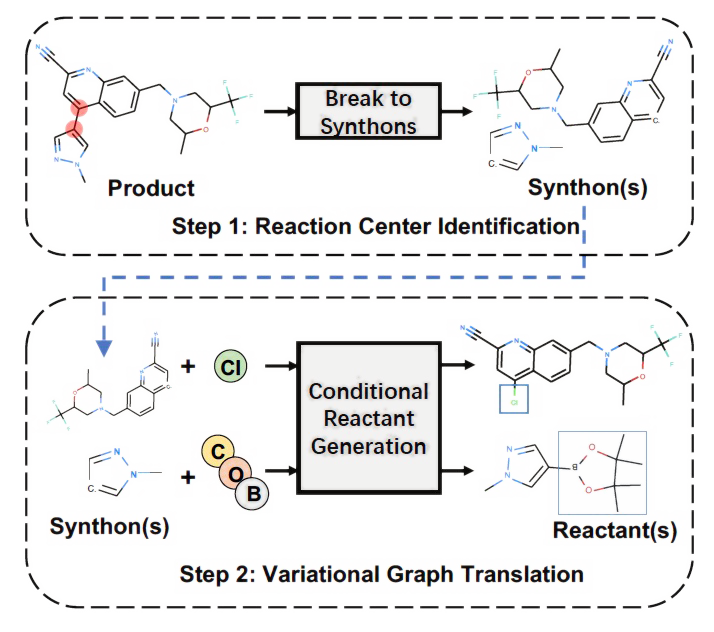
G2G: A Graph to Graphs Framework for Retrosynthesis Prediction
Retrosynthesis via graph translation; inspired by the disconnection approach in Organic Synthesis.
ICML 2020
