BioGeometry, an AI macromolecular pharmaceutical company that just announced the closing of a tens-of-million-dollar level round of exclusive funding from GaoRong Capital in September this year, announces a breakthrough in antibody drug development.
Founded in 2021 by Dr. Jian Tang, an associate professor and tenured professor at the University of Montreal’s Institute of Algorithm Research (Mila), the company, together with Yanling Wu’s team at the Shanghai Engineering Research Center for Synthetic Immunology, has made significant progress in antibody optimization. Through closed-loop dry and wet experiments, BioGeometry’s antibody design platform multiplied the affinity of antibodies, verifying its reliability and efficiency, and opening up a new direction for future antibody drug development.
The Power of Geometric Pretraining
The continuous growth of the antibody drug market has led to an increasing interest in the development of pharmaceutical antibodies.
With the development of genetic engineering and antibody display technology, antibody candidates can be easily obtained through library construction and screening. However, antibodies obtained through this procedure have not undergone in vivo affinity maturation, hence often suffer from inadequate affinity levels, preventing them from therapeutic use. In these cases, in vitro affinity maturation is often required.
In vitro antibody affinity maturation is mainly done by mimicking the in vivo process, adopting various strategies to mutate antibody genes accordingly, constructing mutant antibody libraries, and obtaining high-affinity antibodies through affinity screening. In the process of in vitro affinity maturation of genetically engineered antibodies, the selection of mutation regions and mutations targets are of key importance. Current mutation strategies can be divided into three major categories, including random mutation, substitution and directed mutation.
However, these traditional affinity maturation strategies are far from perfect. “Traditional affinity maturation protocols rely on extensive rounds of experiments including mutation, display and screening. The time for each cycle could take more than two months. Long cycle time, high cost and low success rate are the drawbacks of traditional protocols,” says Dr. Jian Tang, founder and CEO of BioGeometry.
Recognizing the above limitations, the industry has evolved to further enhance the affinity maturation efficacy by adopting different approaches, such as combining various mutation strategies based on antibody characteristics and creating a synergistic effect.
Meanwhile, the advancements in computer-aided drug design and the enrichment of available antibody structures have made it possible to achieve computer-aided directed affinity maturation based on the knowledge mined from antibody structures. Its core advantage lies in its cost-effectiveness, allowing scientists to get improved mutants or better understand the antibody-antigen interaction in a short period of time.
Existing structure prediction models are empowering the above effort. They are met with some success but also limitations. “Structure prediction models such as AlphaFold2 are insensitive to mutations and are particularly inaccurate for modeling protein side chain atoms at the binding interface.” In response to the problems of existing structure prediction models, BioGeometry developed its own atomic-level geometric deep learning model and pretrained it on protein complex structure data. The model enjoys fast and effective modeling of antigen-antibody interactions, achieving new state-of-the-art in the task of mutant affinity prediction.
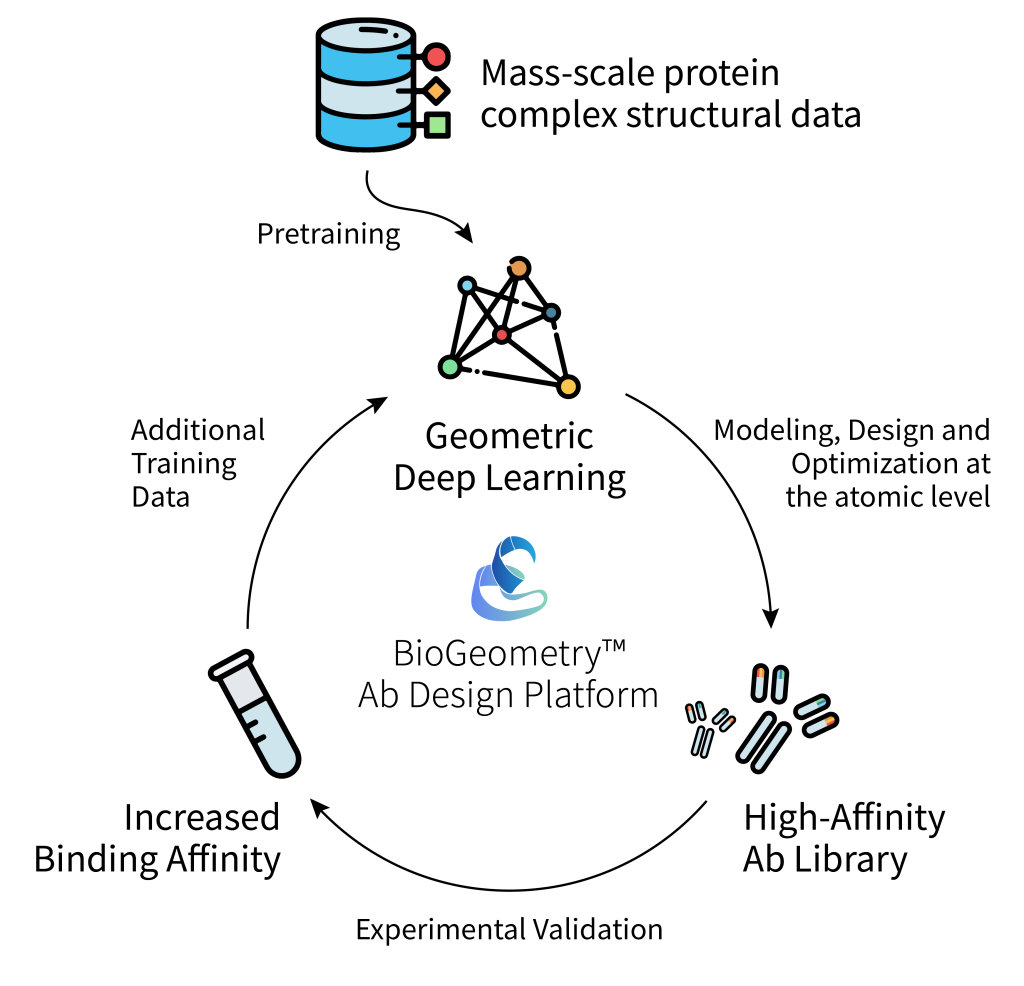
“Pretrained large models have made huge breakthroughs in areas such as images, natural language and protein sequences, but how to pretrain on the 3D structures of proteins and their complexes remains a difficult and hot spot. Our work combines dry and wet experiments and demonstrates the prominent role of pretrained geometric deep learning models in the field of antibody optimization.” commented Dr. Jian Tang.
It is especially worth mentioning that the inference speed of geometric deep learning models is hundreds or even thousands of times faster than traditional molecular dynamics methods, and thus makes large-scale virtual screening and multi-target antibody design possible. This would likely lead to time cost savings, economic benefits and improved success rates in the design of future pharmaceutical antibody leads.
Ultimately, the high-affinity antibodies would further reduce required doses and subsequent toxic reactions or side effects, and enhance therapeutic efficacy.
Towards a One-Stop Antibody Design Platform
Previously, the BioGeometry team, together with Nvidia, Intel, IBM and other companies, released TorchProtein, the first open-source machine learning platform for macromolecular drug discovery. The platform also includes the first pretrained big model for protein 3D geometry and a standard dataset for evaluating the effectiveness of deep learning on protein modeling.
Based on its technology repertoire, BioGeometry has completed the construction of its AI macromolecule drug design platform. Now, it is pushing deeper into the field, building a one-stop antibody design platform based on pretrained geometric deep learning models, which can optimize affinity, together with other important developability properties such as stability, solubility and viscosity simultaneously to accelerate the antibody development process.
According to Dr. Tang, traditionally, affinity and the various developability properties are optimized incrementally rather than simultaneously. This may lead to problems such as a decrease in stability while improving affinity. The adoption of AI opens up wild possibilities towards developing much more balanced pharmaceutical antibody leads in terms of such properties.
In practice, pharmaceutical antibody leads often run into immunogenicity problems. Immunogenicity refers to the ability of a drug to provoke an immune response or related events in the patient body against the drug itself or related proteins, inducing adverse events including the formation of anti-drug antibodies. These might lead to strong, sometimes life-threatening, immune responses in patients; and/or neutralization of the biological activity and thus efficacy of the drug.
In order to better address these issues, immunogenicity has been taken into account in BioGeometry’s one-stop antibody design platform. The platform will try to improve the homology between the antibody sequence and the human ones. The risk of immunogenicity could be further reduced by predicting the risk of immunogenicity with AI, followed by targeted removal of high-risk sites through protein engineering.
Dr. Tang said that affinity optimization is essentially a part of antibody optimization. In the future, BioGeometry’s goal is to design antibodies entirely by computers. Armed with an AI macromolecule drug design platform, BioGeometry now dives deep with with its one-stop antibody design platform. It is also conducting more cutting-edge explorations around antibody structure prediction, antibody optimization, antibody sequence design, enzyme activity prediction and other fields where the team is already taking a leading position. Moreover, while further expanding its internal product pipelines, BioGeometry also aims to further commercialize, e.g. helping pharmaceutical companies to optimize their antibodies using its one-stop antibody design platform.
With its collaboration with several academic institutions and pharmaceutical companies pushes forward, BioGeometry is actively accelerating its technology development, verification and deployment to advance the drug candidates to the clinical stage in the near future.
